KOMPUTE method testing - BC data (v10.1)
Coby Warkentin and Donghyung Lee
2023-06-19
- Load packages
- Prep control phenotype data
- Load Body Composition control phenotypes
- Heatmap showing measured phenotypes
- Remove phenotypes with num of obs < 15000
- Remove samples with num of measured phenotypes < 7
- Heapmap of measured phenotypes after filtering
- Reshape the data (long to wide)
- Distribution of each phenotype
- Rank Z transformation
- Principal Variance Component Analysis
- Removing batch effects using ComBat
- PVCA on residuals from ComBat
- Compute correlations between phenotypes
- Prep IMPC summary stat
- Read BC summary stat (IMPCv10.1)
- Duplicates in gene-phenotype pair
- Using Stouffer’s method, merge multiple z-scores of a gene-phenotype pair into a z-score
- Make z-score matrix (long to wide)
- Z-score Distribution
- Estimate genetic correlation matrix between phenotypes using Zscores
- Phenotype Corr VS Genetic Corr btw phenotypes
- Test of the correlation between genetic correlation matrices
- Test KOMPUTE imputation algorithm
Last updated: 2023-06-19
Checks: 7 0
Knit directory: komputeExamples/
This reproducible R Markdown analysis was created with workflowr (version 1.7.0.1). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20230110) was run prior to running
the code in the R Markdown file. Setting a seed ensures that any results
that rely on randomness, e.g. subsampling or permutations, are
reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility.
The results in this page were generated with repository version bb7f5ee. See the Past versions tab to see a history of the changes made to the R Markdown and HTML files.
Note that you need to be careful to ensure that all relevant files for
the analysis have been committed to Git prior to generating the results
(you can use wflow_publish or
wflow_git_commit). workflowr only checks the R Markdown
file, but you know if there are other scripts or data files that it
depends on. Below is the status of the Git repository when the results
were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: analysis/.DS_Store
Ignored: code/.DS_Store
Untracked files:
Untracked: analysis/figures.Rmd_old
Unstaged changes:
Deleted: analysis/figures.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the repository in which changes were
made to the R Markdown (analysis/kompute_test_BC_v10.1.Rmd)
and HTML (docs/kompute_test_BC_v10.1.html) files. If you’ve
configured a remote Git repository (see ?wflow_git_remote),
click on the hyperlinks in the table below to view the files as they
were in that past version.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| html | 7685a09 | statsleelab | 2023-01-10 | first commit |
| Rmd | 32dd775 | statsleelab | 2023-01-10 | first commit |
Load packages
rm(list=ls())
library(data.table)
library(dplyr)
library(reshape2)
library(ggplot2)
library(tidyr) #spread
library(RColorBrewer)
#library(irlba) # partial PCA
#library(cowplot)
library(circlize)
library(ComplexHeatmap)Prep control phenotype data
Load Body Composition control phenotypes
BC.data <- readRDS("data/BC.data.rds")
dim(BC.data)[1] 243532 10Heatmap showing measured phenotypes
This heatmaps show phenotypes measured for each control mouse. Columns represent mice and rows represent phenotypes.
mtest <- table(BC.data$proc_param_name_stable_id, BC.data$biological_sample_id)
mtest <-as.data.frame.matrix(mtest)
dim(mtest)[1] 51 25690if(FALSE){
nmax <-max(mtest)
library(circlize)
col_fun = colorRamp2(c(0, nmax), c("white", "red"))
col_fun(seq(0, nmax))
ht = Heatmap(as.matrix(mtest), cluster_rows = FALSE, cluster_columns = FALSE, show_column_names = F, col = col_fun,
row_names_gp = gpar(fontsize = 8), name="Count")
draw(ht)
}Remove phenotypes with num of obs < 15000
mtest <- table(BC.data$proc_param_name, BC.data$biological_sample_id)
dim(mtest)[1] 10 25690#head(mtest[,1:10])
mtest0 <- mtest>0
#head(mtest0[,1:10])
rowSums(mtest0) BC_BMC/Body weight
24446
BC_Body length
19233
BC_Body weight
25675
BC_Bone Area
24461
BC_Bone Mineral Content (excluding skull)
24714
BC_Bone Mineral Density (excluding skull)
24715
BC_Fat mass
24950
BC_Fat/Body weight
24682
BC_Lean mass
24952
BC_Lean/Body weight
24684 rmv.pheno.list <- rownames(mtest)[rowSums(mtest0)<15000]
#rmv.pheno.list
dim(BC.data)[1] 243532 10BC.data <- BC.data %>% filter(!(proc_param_name %in% rmv.pheno.list))
dim(BC.data)[1] 243532 10# number of phenotypes left
length(unique(BC.data$proc_param_name))[1] 10Remove samples with num of measured phenotypes < 7
mtest <- table(BC.data$proc_param_name, BC.data$biological_sample_id)
dim(mtest)[1] 10 25690head(mtest[,1:10])
39638 39639 39640 39641 39642 39643
BC_BMC/Body weight 1 1 1 1 1 1
BC_Body length 0 0 0 1 0 1
BC_Body weight 2 1 1 1 1 1
BC_Bone Area 1 1 1 1 1 1
BC_Bone Mineral Content (excluding skull) 2 1 1 1 1 1
BC_Bone Mineral Density (excluding skull) 2 1 1 1 1 1
39650 39651 39652 39657
BC_BMC/Body weight 1 1 1 1
BC_Body length 1 0 0 1
BC_Body weight 1 1 1 1
BC_Bone Area 1 1 1 1
BC_Bone Mineral Content (excluding skull) 1 1 1 1
BC_Bone Mineral Density (excluding skull) 1 1 1 1mtest0 <- mtest>0
head(mtest0[,1:10])
39638 39639 39640 39641 39642 39643
BC_BMC/Body weight TRUE TRUE TRUE TRUE TRUE TRUE
BC_Body length FALSE FALSE FALSE TRUE FALSE TRUE
BC_Body weight TRUE TRUE TRUE TRUE TRUE TRUE
BC_Bone Area TRUE TRUE TRUE TRUE TRUE TRUE
BC_Bone Mineral Content (excluding skull) TRUE TRUE TRUE TRUE TRUE TRUE
BC_Bone Mineral Density (excluding skull) TRUE TRUE TRUE TRUE TRUE TRUE
39650 39651 39652 39657
BC_BMC/Body weight TRUE TRUE TRUE TRUE
BC_Body length TRUE FALSE FALSE TRUE
BC_Body weight TRUE TRUE TRUE TRUE
BC_Bone Area TRUE TRUE TRUE TRUE
BC_Bone Mineral Content (excluding skull) TRUE TRUE TRUE TRUE
BC_Bone Mineral Density (excluding skull) TRUE TRUE TRUE TRUEsummary(colSums(mtest0)) Min. 1st Qu. Median Mean 3rd Qu. Max.
1.00 9.00 10.00 9.44 10.00 10.00 rmv.sample.list <- colnames(mtest)[colSums(mtest0)<7]
length(rmv.sample.list)[1] 1829dim(BC.data)[1] 243532 10BC.data <- BC.data %>% filter(!(biological_sample_id %in% rmv.sample.list))
dim(BC.data)[1] 234305 10# number of observations to use
length(unique(BC.data$biological_sample_id))[1] 23861Heapmap of measured phenotypes after filtering
if(FALSE){
mtest <- table(BC.data$proc_param_name, BC.data$biological_sample_id)
dim(mtest)
mtest <-as.data.frame.matrix(mtest)
nmax <-max(mtest)
library(circlize)
col_fun = colorRamp2(c(0, nmax), c("white", "red"))
col_fun(seq(0, nmax))
pdf("~/Google Drive Miami/Miami_IMPC/output/measured_phenotypes_controls_after_filtering_BC.pdf", width = 10, height = 3)
ht = Heatmap(as.matrix(mtest), cluster_rows = FALSE, cluster_columns = FALSE, show_column_names = F, col = col_fun,
row_names_gp = gpar(fontsize = 7), name="Count")
draw(ht)
dev.off()
}Reshape the data (long to wide)
BC.mat <- BC.data %>%
dplyr::select(biological_sample_id, proc_param_name, data_point, sex, phenotyping_center, strain_name) %>%
##consider weight or age in weeks
arrange(biological_sample_id) %>%
distinct(biological_sample_id, proc_param_name, .keep_all=TRUE) %>% ## remove duplicates, maybe mean() is better.
spread(proc_param_name, data_point) %>%
tibble::column_to_rownames(var="biological_sample_id")
head(BC.mat) sex phenotyping_center strain_name BC_BMC/Body weight BC_Body length
39638 female MRC Harwell C57BL/6NTac 0.0567631 NA
39639 female HMGU C57BL/6NCrl 0.0224897 NA
39640 female HMGU C57BL/6NTac 0.0214276 NA
39641 male HMGU C57BL/6NCrl 0.0191929 9.7
39642 female MRC Harwell C57BL/6NTac 0.0242145 NA
39643 female HMGU C57BL/6NCrl 0.0224004 9.4
BC_Body weight BC_Bone Area BC_Bone Mineral Content (excluding skull)
39638 22.12 13.51560 1.25560
39639 23.30 8.11161 0.52401
39640 23.20 6.85683 0.49712
39641 29.70 8.61072 0.57003
39642 25.27 8.42837 0.61190
39643 24.30 8.42616 0.54433
BC_Bone Mineral Density (excluding skull) BC_Fat mass BC_Fat/Body weight
39638 0.0929 6.9362 0.313571
39639 0.0646 6.1000 0.261803
39640 0.0725 3.3000 0.142241
39641 0.0662 7.1000 0.239057
39642 0.0726 3.4382 0.136059
39643 0.0646 6.9000 0.283951
BC_Lean mass BC_Lean/Body weight
39638 11.2569 0.508901
39639 14.7000 0.630901
39640 17.1000 0.737069
39641 21.7000 0.730640
39642 20.5392 0.812790
39643 15.3000 0.629630dim(BC.mat)[1] 23861 13summary(colSums(is.na(BC.mat[,-1:-3]))) Min. 1st Qu. Median Mean 3rd Qu. Max.
0.00 0.00 0.00 532.50 0.75 5323.00 Distribution of each phenotype
ggplot(melt(BC.mat), aes(x=value)) +
geom_histogram() +
facet_wrap(~variable, scales="free", ncol=5)+
theme(strip.text.x = element_text(size = 6))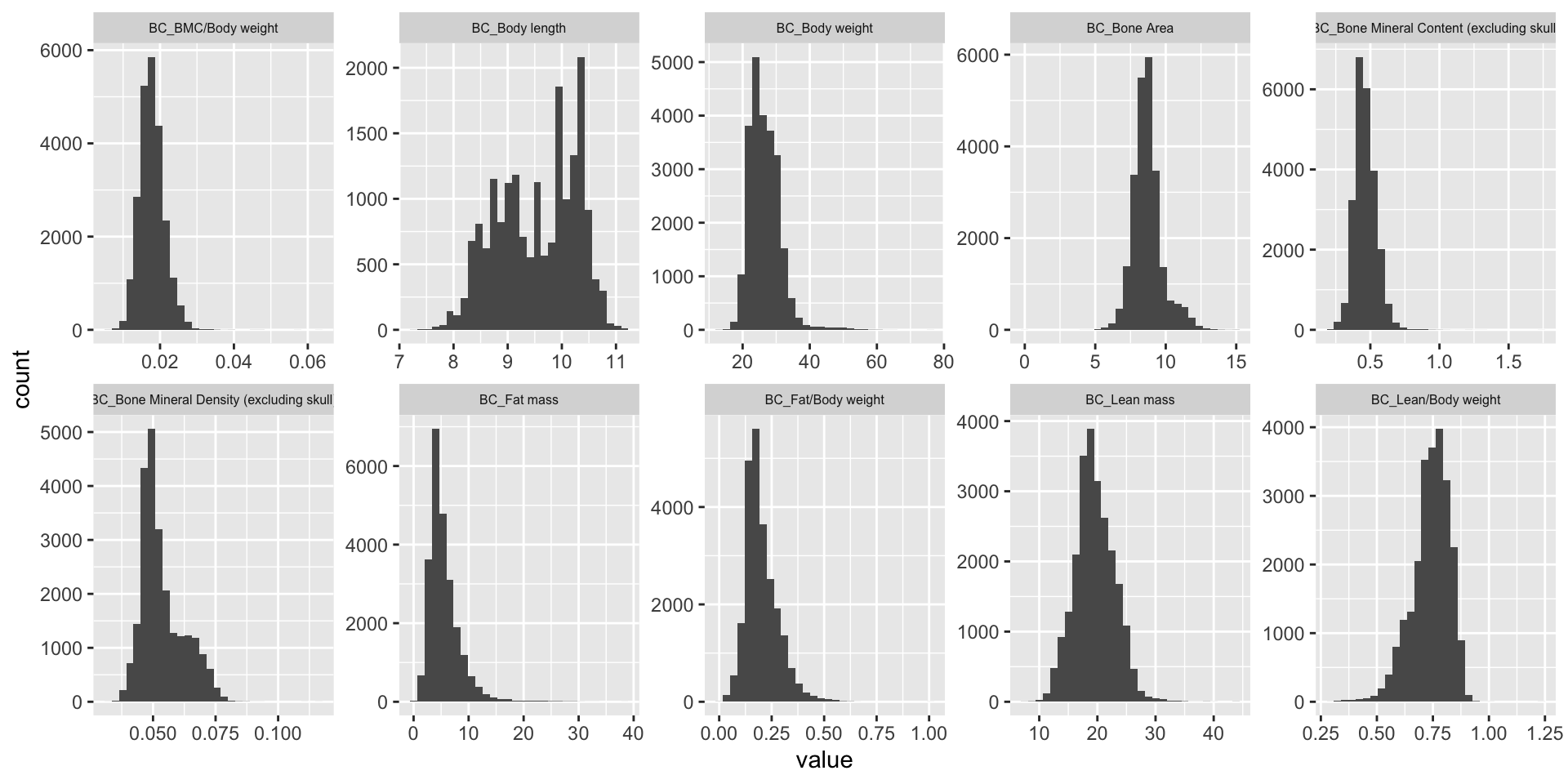
| Version | Author | Date |
|---|---|---|
| 7685a09 | statsleelab | 2023-01-10 |
Rank Z transformation
library(RNOmni)
BC.mat.rank <- BC.mat
dim(BC.mat.rank)[1] 23861 13BC.mat.rank <- BC.mat.rank[complete.cases(BC.mat.rank),]
dim(BC.mat.rank)[1] 18538 13dim(BC.mat)[1] 23861 13BC.mat <- BC.mat[complete.cases(BC.mat),]
dim(BC.mat)[1] 18538 13BC.mat.rank <- cbind(BC.mat.rank[,1:3], apply(BC.mat.rank[,-1:-3], 2, RankNorm))
ggplot(melt(BC.mat.rank), aes(x=value)) +
geom_histogram() +
facet_wrap(~variable, scales="free", ncol=5)+
theme(strip.text.x = element_text(size = 6))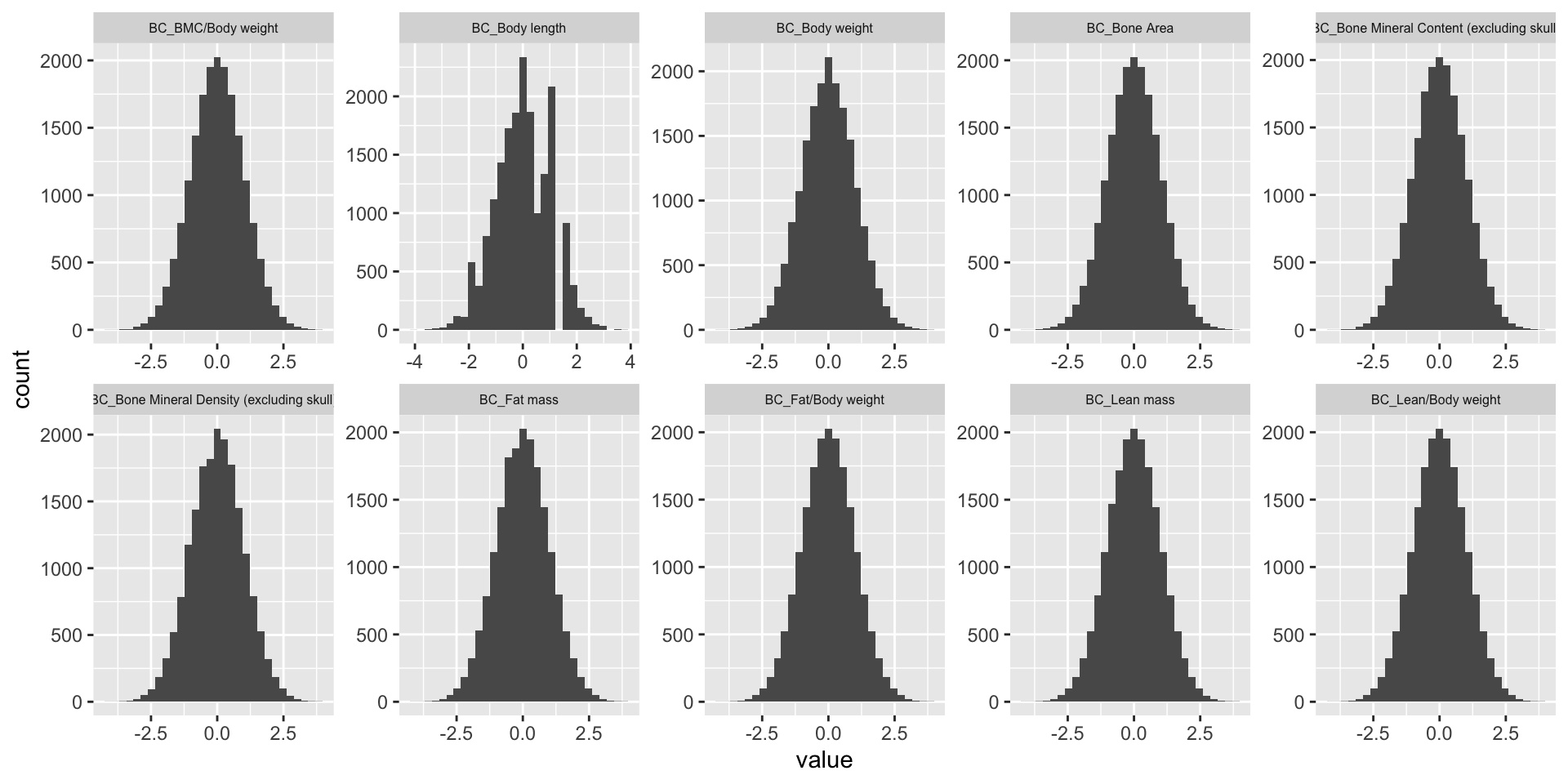
| Version | Author | Date |
|---|---|---|
| 7685a09 | statsleelab | 2023-01-10 |
Principal Variance Component Analysis
Here we conducted a PVCA analysis on the phenotype matrix data and measure the proportion of variance explained by each important covariate (sex, phenotyping_center).
source("code/PVCA.R")
meta <- BC.mat.rank[,1:3] ## looking at covariates sex, phenotyping_center, and strain_name
head(meta) sex phenotyping_center strain_name
39641 male HMGU C57BL/6NCrl
39643 female HMGU C57BL/6NCrl
39657 male HMGU C57BL/6NCrl
39750 female HMGU C57BL/6NCrl
39763 female HMGU C57BL/6NCrl
39773 female HMGU C57BL/6NCrldim(meta)[1] 18538 3summary(meta) # variables are still characters sex phenotyping_center strain_name
Length:18538 Length:18538 Length:18538
Class :character Class :character Class :character
Mode :character Mode :character Mode :character meta[sapply(meta, is.character)] <- lapply(meta[sapply(meta, is.character)], as.factor)
summary(meta) # now all variables are converted to factors sex phenotyping_center strain_name
female:9282 WTSI :4573 B6Brd;B6Dnk;B6N-Tyr<c-Brd>: 176
male :9256 JAX :3675 C57BL/6N :9127
ICS :2075 C57BL/6NCrl :2997
BCM :1832 C57BL/6NJ :3675
RBRC :1692 C57BL/6NJcl : 728
UC Davis:1441 C57BL/6NTac :1835
(Other) :3250 chisq.test(meta[,1],meta[,2])
Pearson's Chi-squared test
data: meta[, 1] and meta[, 2]
X-squared = 13.572, df = 10, p-value = 0.1934chisq.test(meta[,2],meta[,3])
Pearson's Chi-squared test
data: meta[, 2] and meta[, 3]
X-squared = 59770, df = 50, p-value < 2.2e-16meta<-meta[,-3] # phenotyping_center and strain_name strongly associated and this caused confouding in PVCA analysis so strain_name dropped.
G <- t(BC.mat.rank[,-1:-3]) ## phenotype matrix data
set.seed(09302021)
# Perform PVCA for 10 random samples of size 1000 (more computationally efficient)
pvca.res <- matrix(nrow=10, ncol=3)
for (i in 1:10){
sample <- sample(1:ncol(G), 1000, replace=FALSE)
pvca.res[i,] <- PVCA(G[,sample], meta[sample,], threshold=0.6, inter=FALSE)
}
# Average effect size across samples
pvca.means <- colMeans(pvca.res)
names(pvca.means) <- c(colnames(meta), "resid")
# Plot PVCA
pvca.plot <- PlotPVCA(pvca.means, "PVCA of Phenotype Matrix Data")
pvca.plot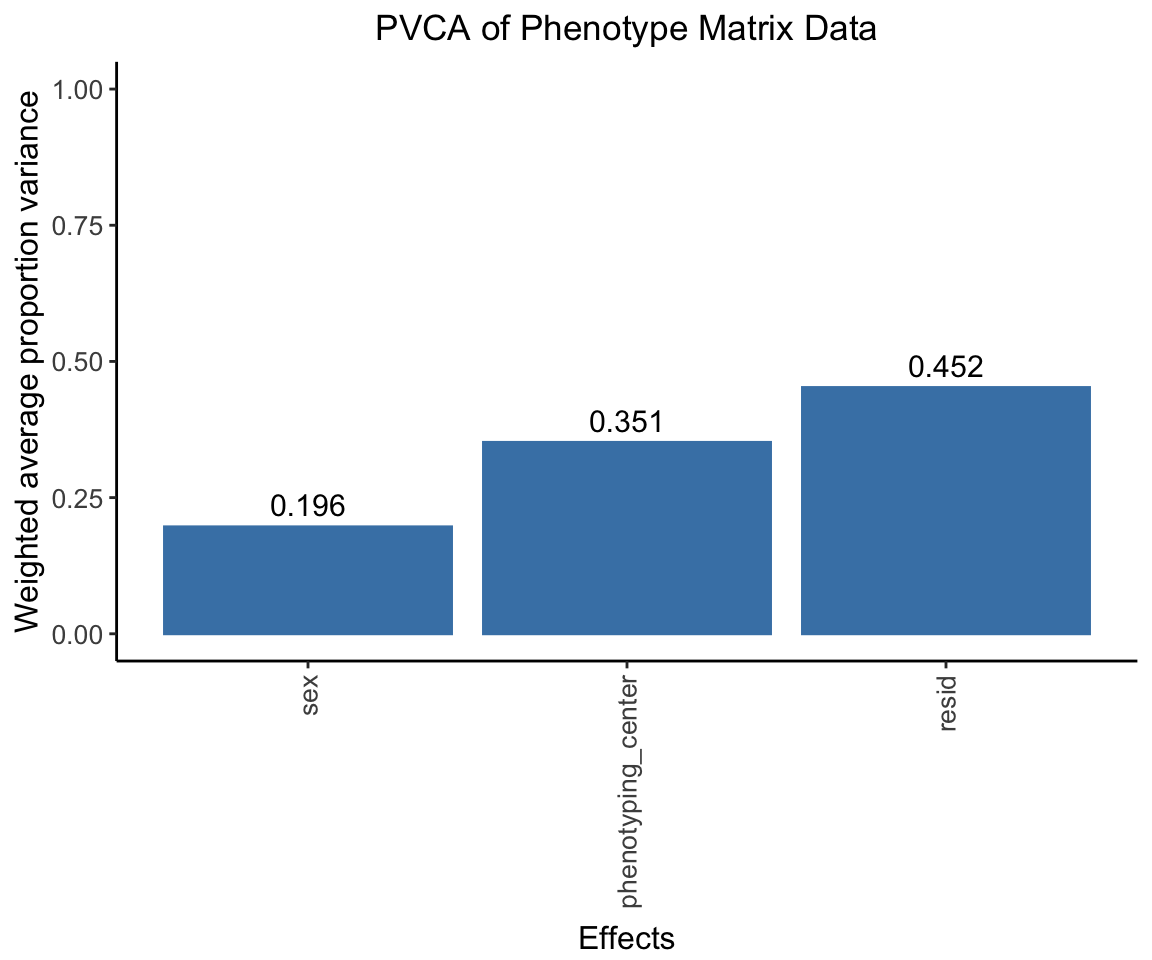
| Version | Author | Date |
|---|---|---|
| 7685a09 | statsleelab | 2023-01-10 |
png(file="docs/figure/figures.Rmd/pvca_BC_1.png", width=600, height=350)
pvca.plot
dev.off()quartz_off_screen
2 Removing batch effects using ComBat
We remove the center effect using ComBat.
library(sva)Loading required package: mgcvLoading required package: nlme
Attaching package: 'nlme'The following object is masked from 'package:lme4':
lmListThe following object is masked from 'package:dplyr':
collapseThis is mgcv 1.8-40. For overview type 'help("mgcv-package")'.Loading required package: genefilter
Attaching package: 'genefilter'The following object is masked from 'package:ComplexHeatmap':
dist2Loading required package: BiocParallelcombat_komp = ComBat(dat=G, batch=meta$phenotyping_center, par.prior=TRUE, prior.plots=TRUE, mod=NULL)Found11batchesAdjusting for0covariate(s) or covariate level(s)Standardizing Data across genesFitting L/S model and finding priorsFinding parametric adjustmentsAdjusting the Data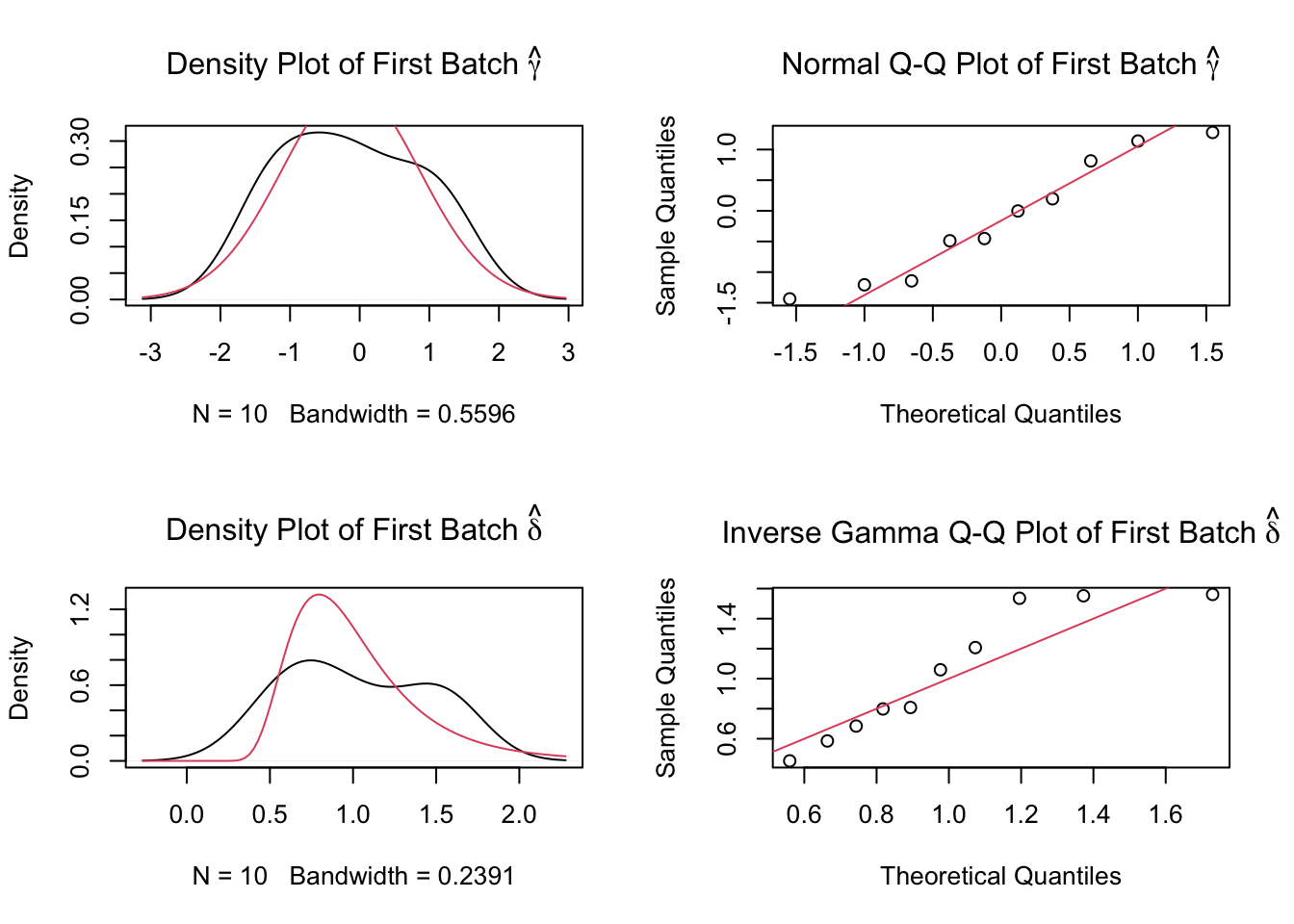
| Version | Author | Date |
|---|---|---|
| 7685a09 | statsleelab | 2023-01-10 |
combat_komp[1:5,1:5] 39641 39643 39657
BC_BMC/Body weight -0.5446577 0.7624261 -0.8478037
BC_Body length 0.2371464 -0.1563519 0.2371464
BC_Body weight 0.9069478 -0.3865968 0.9844317
BC_Bone Area 0.4188173 0.1510426 1.0646332
BC_Bone Mineral Content (excluding skull) 0.5872088 0.1561049 0.3622582
39750 39763
BC_BMC/Body weight -0.06815544 0.14454428
BC_Body length -0.28891231 -0.01379022
BC_Body weight -0.59375686 -0.59375686
BC_Bone Area -0.51719766 -0.29607359
BC_Bone Mineral Content (excluding skull) -0.84928634 -0.64311406G[1:5,1:5] # for comparison, combat_komp is same form and same dimensions as G 39641 39643 39657
BC_BMC/Body weight 0.61537676 1.3908581 0.43552296
BC_Body length 0.04917004 -0.1811585 0.04917004
BC_Body weight 0.72940618 -0.3554534 0.79438976
BC_Bone Area -0.05160698 -0.2731793 0.48277848
BC_Bone Mineral Content (excluding skull) 1.43981495 1.1313384 1.27885140
39750 39763
BC_BMC/Body weight 0.8980814 1.02427422
BC_Body length -0.2587508 -0.09771206
BC_Body weight -0.5291927 -0.52919275
BC_Bone Area -0.8261201 -0.64314918
BC_Bone Mineral Content (excluding skull) 0.4119303 0.55945691PVCA on residuals from ComBat
The center effect should be much lower.
set.seed(09302021)
# Perform PVCA for 10 samples (more computationally efficient)
pvca.res.nobatch <- matrix(nrow=10, ncol=3)
for (i in 1:10){
sample <- sample(1:ncol(combat_komp), 1000, replace=FALSE)
pvca.res.nobatch[i,] <- PVCA(combat_komp[,sample], meta[sample,], threshold=0.6, inter=FALSE)
}
# Average effect size across samples
pvca.means.nobatch <- colMeans(pvca.res.nobatch)
names(pvca.means.nobatch) <- c(colnames(meta), "resid")
# Plot PVCA
pvca.plot.nobatch <- PlotPVCA(pvca.means.nobatch, "PVCA of Phenotype Matrix Data with Reduced Batch Effect")
pvca.plot.nobatch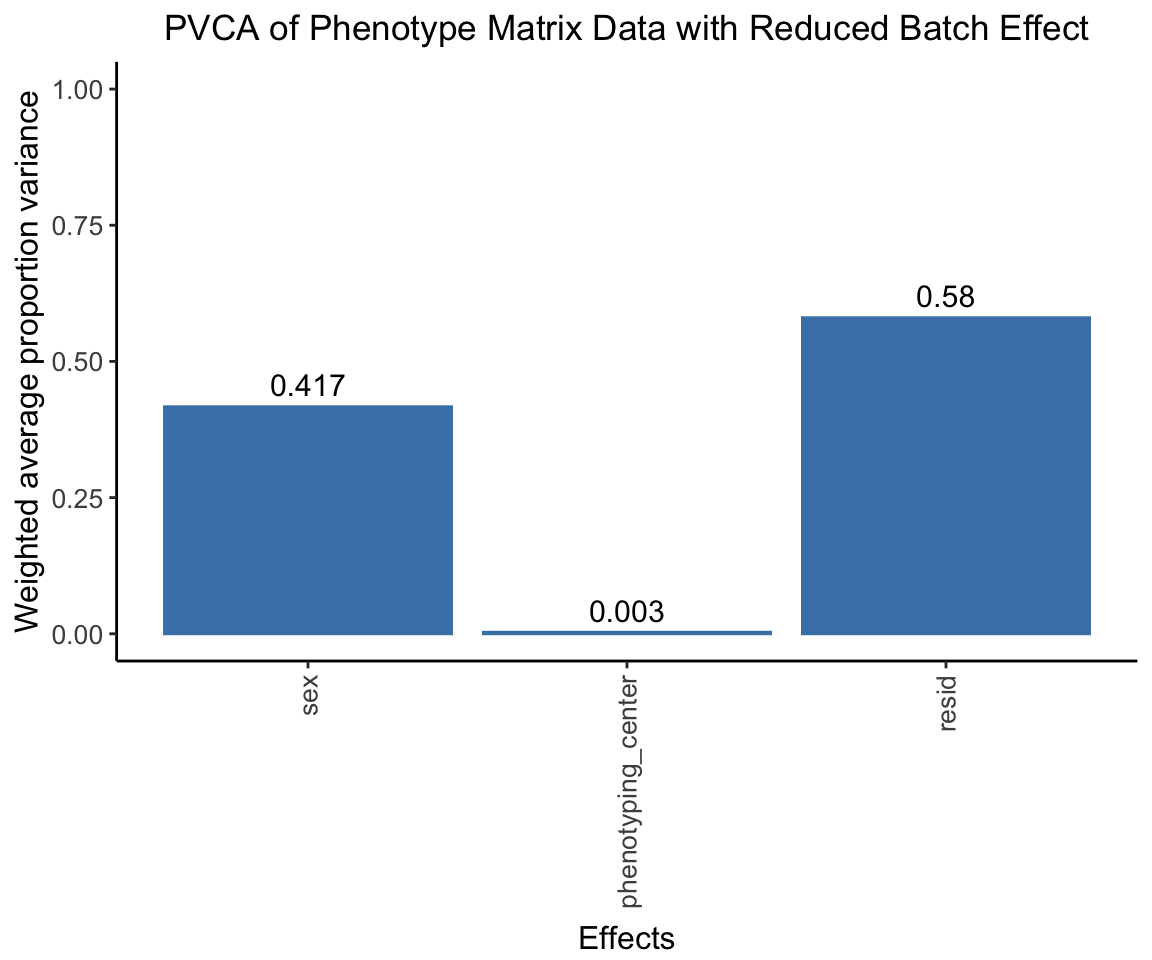
| Version | Author | Date |
|---|---|---|
| 7685a09 | statsleelab | 2023-01-10 |
png(file="docs/figure/figures.Rmd/pvca_BC_2.png", width=600, height=350)
pvca.plot.nobatch
dev.off()quartz_off_screen
2 Compute correlations between phenotypes
BC.cor.rank <- cor(BC.mat.rank[,-1:-3], use="pairwise.complete.obs") # pearson correlation coefficient
BC.cor <- cor(BC.mat[,-1:-3], use="pairwise.complete.obs", method="spearman") # spearman
BC.cor.combat <- cor(t(combat_komp), use="pairwise.complete.obs")
pheno.list <- rownames(BC.cor)
ht1 = Heatmap(BC.cor, show_column_names = F, row_names_gp = gpar(fontsize = 9), name="Spearm. Corr.")
draw(ht1)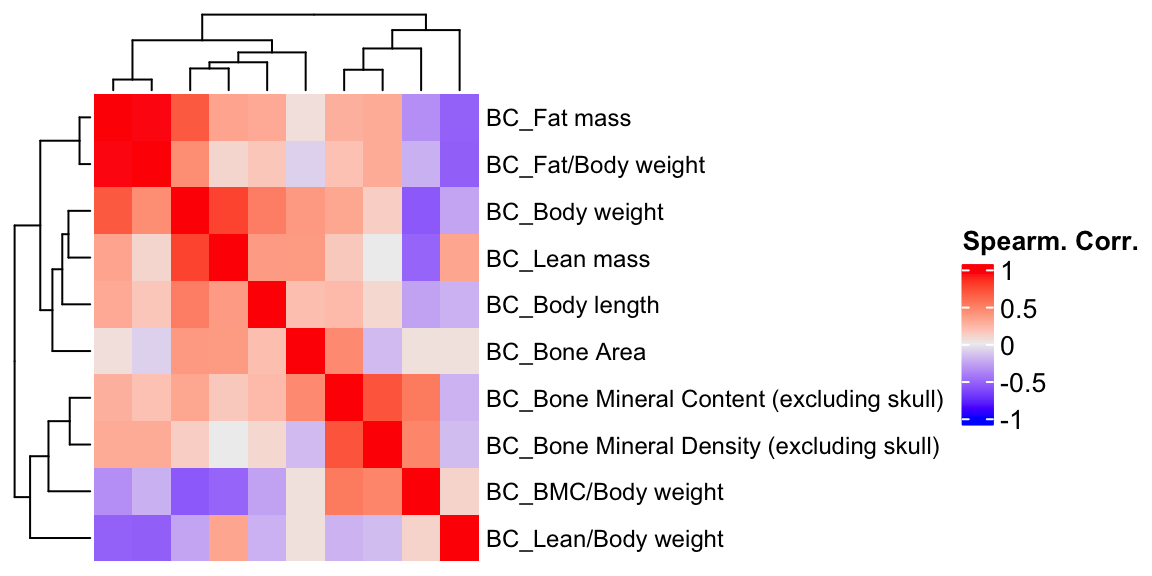
| Version | Author | Date |
|---|---|---|
| 7685a09 | statsleelab | 2023-01-10 |
ht2 = Heatmap(BC.cor.rank, show_column_names = F, row_names_gp = gpar(fontsize = 9), name="Corr. RankZ")
draw(ht2)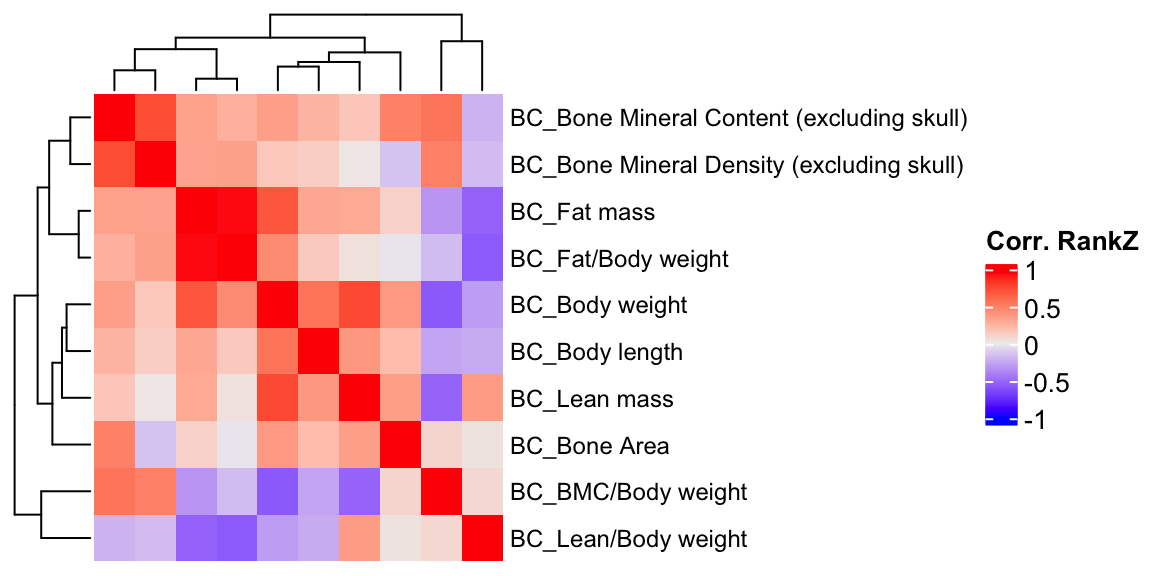
| Version | Author | Date |
|---|---|---|
| 7685a09 | statsleelab | 2023-01-10 |
ht3 = Heatmap(BC.cor.combat, show_column_names = F, row_names_gp = gpar(fontsize = 9), name="Corr. ComBat")
draw(ht3)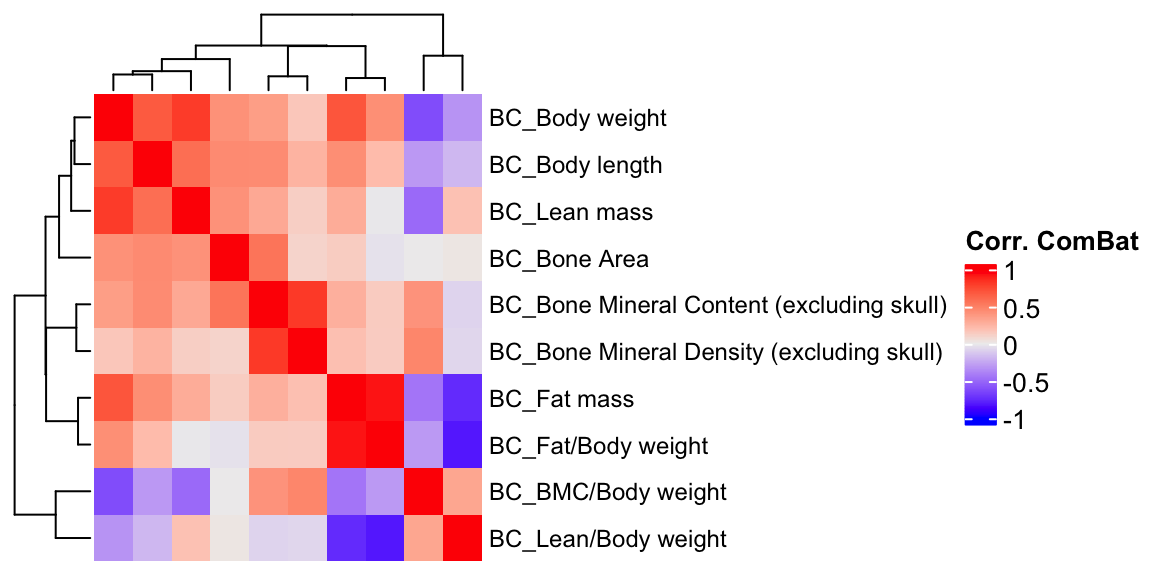
| Version | Author | Date |
|---|---|---|
| 7685a09 | statsleelab | 2023-01-10 |
Prep IMPC summary stat
Read BC summary stat (IMPCv10.1)
BC.stat <- readRDS("data/BC.stat.rds")
dim(BC.stat)[1] 42582 8table(BC.stat$parameter_name, BC.stat$procedure_name)
BC
BMC/Body weight 4780
Body length 4197
Bone Area 4780
Bone Mineral Content (excluding skull) 4780
Bone Mineral Density (excluding skull) 4780
Fat mass 4816
Fat/Body weight 4815
Lean mass 4817
Lean/Body weight 4817length(unique(BC.stat$marker_symbol)) #3362[1] 4428length(unique(BC.stat$allele_symbol)) #3412[1] 4559length(unique(BC.stat$proc_param_name)) #15 # number of phenotypes in association statistics data set[1] 9length(unique(BC.data$proc_param_name)) #15 # number of phenotypes in final control data[1] 10pheno.list.stat <- unique(BC.stat$proc_param_name)
pheno.list.ctrl <- unique(BC.data$proc_param_name)
sum(pheno.list.stat %in% pheno.list.ctrl)[1] 9sum(pheno.list.ctrl %in% pheno.list.stat)[1] 9## extract common phenotype list
common.pheno.list <- sort(intersect(pheno.list.ctrl, pheno.list.stat))
common.pheno.list[1] "BC_BMC/Body weight"
[2] "BC_Body length"
[3] "BC_Bone Area"
[4] "BC_Bone Mineral Content (excluding skull)"
[5] "BC_Bone Mineral Density (excluding skull)"
[6] "BC_Fat mass"
[7] "BC_Fat/Body weight"
[8] "BC_Lean mass"
[9] "BC_Lean/Body weight" length(common.pheno.list) # 14 - each data set had one phenotype not present in the other[1] 9## Use summary statistics of common phenotypes
dim(BC.stat)[1] 42582 8BC.stat <- BC.stat %>% filter(proc_param_name %in% common.pheno.list)
dim(BC.stat)[1] 42582 8length(unique(BC.stat$proc_param_name))[1] 9Duplicates in gene-phenotype pair
mtest <- table(BC.stat$proc_param_name, BC.stat$marker_symbol)
mtest <-as.data.frame.matrix(mtest)
nmax <-max(mtest)
col_fun = colorRamp2(c(0, nmax), c("white", "red"))
col_fun(seq(0, nmax)) [1] "#FFFFFFFF" "#FFF0EBFF" "#FFE2D7FF" "#FFD3C4FF" "#FFC4B0FF" "#FFB59DFF"
[7] "#FFA68BFF" "#FF9678FF" "#FF8666FF" "#FF7554FF" "#FF6342FF" "#FF4E2FFF"
[13] "#FF351BFF" "#FF0000FF"ht = Heatmap(as.matrix(mtest), cluster_rows = FALSE, cluster_columns = FALSE, show_column_names = F, col = col_fun,
row_names_gp = gpar(fontsize = 8), name="Count")
draw(ht)
| Version | Author | Date |
|---|---|---|
| 7685a09 | statsleelab | 2023-01-10 |
Using Stouffer’s method, merge multiple z-scores of a gene-phenotype pair into a z-score
## sum(z-score)/sqrt(# of zscore)
sumz <- function(z){ sum(z)/sqrt(length(z)) }
BC.z = BC.stat %>%
dplyr::select(marker_symbol, proc_param_name, z_score) %>%
na.omit() %>%
group_by(marker_symbol, proc_param_name) %>%
summarize(zscore = sumz(z_score)) ## combine z-scores`summarise()` has grouped output by 'marker_symbol'. You can override using the
`.groups` argument.dim(BC.z)[1] 32174 3Make z-score matrix (long to wide)
nan2na <- function(df){
out <- data.frame(sapply(df, function(x) ifelse(is.nan(x), NA, x)))
colnames(out) <- colnames(df)
out
}
BC.zmat = dcast(BC.z, marker_symbol ~ proc_param_name, value.var = "zscore",
fun.aggregate = mean) %>% tibble::column_to_rownames(var="marker_symbol")
BC.zmat = nan2na(BC.zmat) #convert nan to na
dim(BC.zmat)[1] 4412 9saveRDS(BC.zmat, file = "data/BC.zmat.rds")
id.mat <- 1*(!is.na(BC.zmat)) # multiply 1 to make this matrix numeric
nrow(as.data.frame(colSums(id.mat)))[1] 9dim(id.mat)[1] 4412 9## heatmap of gene - phenotype (red: tested, white: untested)
ht = Heatmap(t(id.mat),
cluster_rows = T, clustering_distance_rows ="binary",
cluster_columns = T, clustering_distance_columns = "binary",
show_row_dend = F, show_column_dend = F, # do not show dendrogram
show_column_names = F, col = c("white","red"),
row_names_gp = gpar(fontsize = 10), name="Missing")
draw(ht)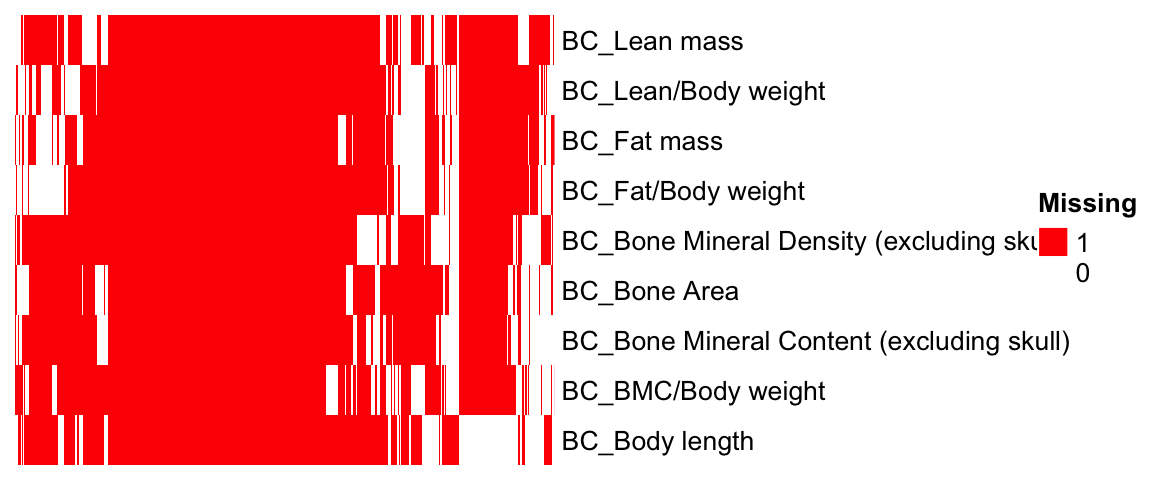
| Version | Author | Date |
|---|---|---|
| 7685a09 | statsleelab | 2023-01-10 |
Z-score Distribution
We plot association Z-score distribution for each phenotype.
ggplot(melt(BC.zmat), aes(x=value)) +
geom_histogram() +
facet_wrap(~variable, scales="free", ncol=5)+
theme(strip.text.x = element_text(size = 6))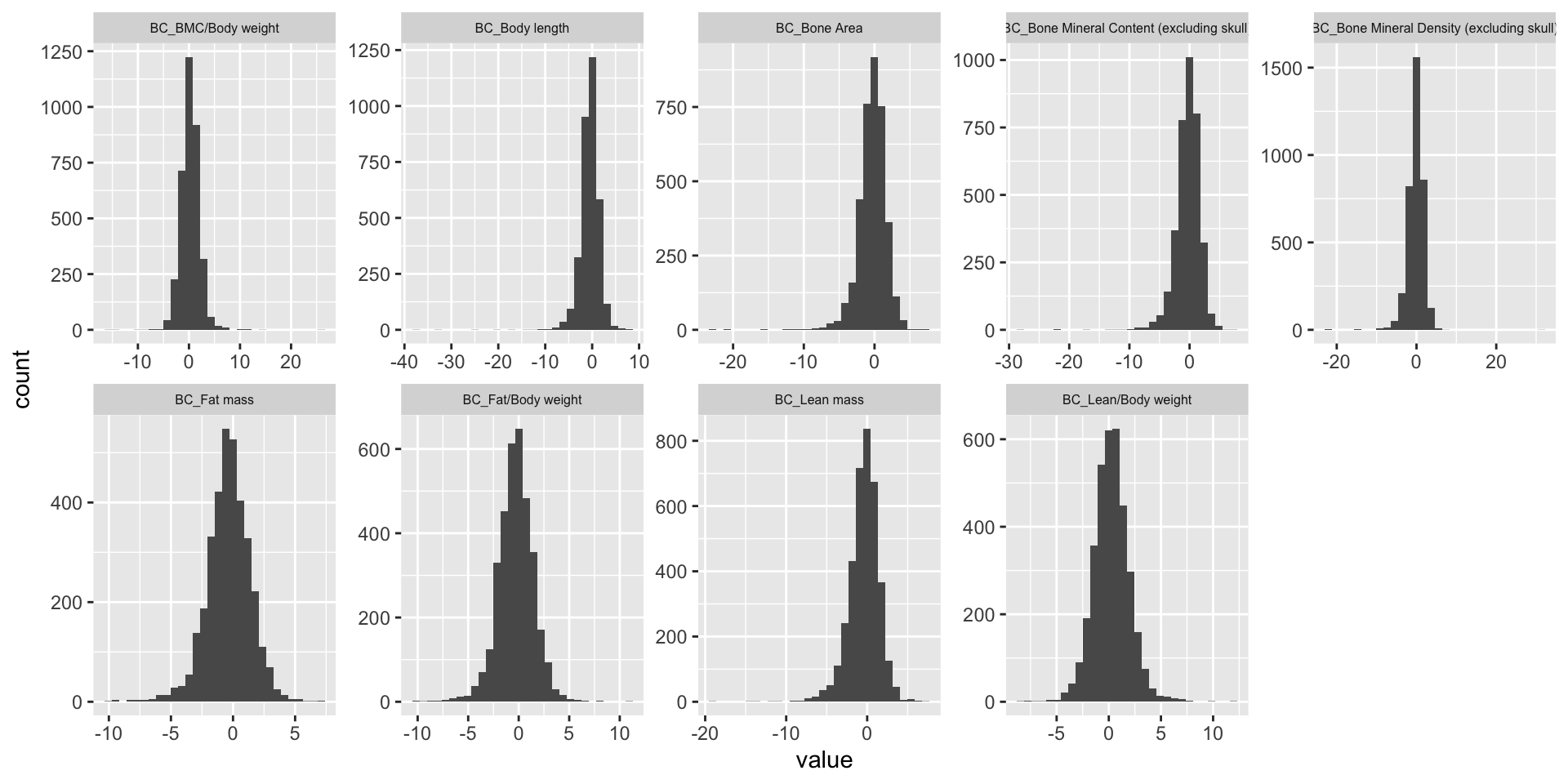
| Version | Author | Date |
|---|---|---|
| 7685a09 | statsleelab | 2023-01-10 |
Estimate genetic correlation matrix between phenotypes using Zscores
Here, we estimate the genetic correlations between phenotypes using association Z-score matrix (num of genes:3983, num of phenotypes 19).
BC.zmat <- BC.zmat[,common.pheno.list]
BC.zcor = cor(BC.zmat, use="pairwise.complete.obs")
ht = Heatmap(BC.zcor, cluster_rows = T, cluster_columns = T, show_column_names = F, #col = col_fun,
row_names_gp = gpar(fontsize = 10),
name="Genetic Corr (Z-score)"
)
draw(ht)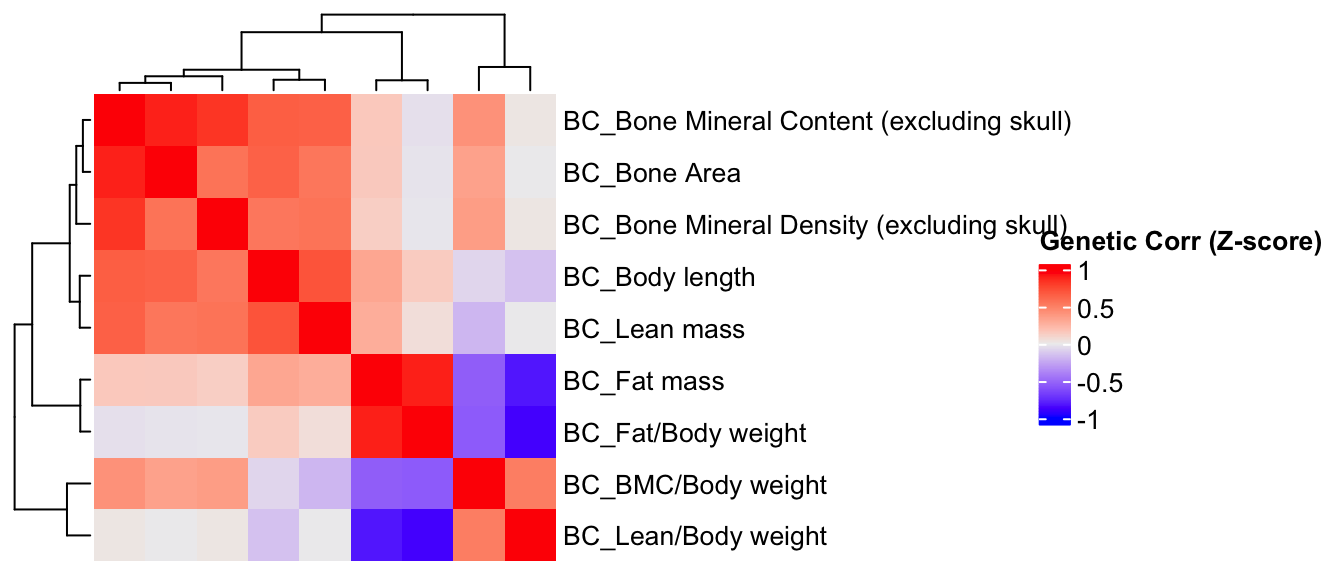
| Version | Author | Date |
|---|---|---|
| 7685a09 | statsleelab | 2023-01-10 |
Phenotype Corr VS Genetic Corr btw phenotypes
We compare a correlation matrix obtained using control mice phenotype data v.s. a genetic correlation matrix estimated using association Z-scores. As you can see, both correlation heatmaps have similar correlation pattern.
BC.cor.rank.fig <- BC.cor.rank[common.pheno.list,common.pheno.list]
BC.cor.fig <- BC.cor[common.pheno.list,common.pheno.list]
BC.cor.combat.fig <- BC.cor.combat[common.pheno.list, common.pheno.list]
BC.zcor.fig <- BC.zcor
ht = Heatmap(BC.cor.rank.fig, cluster_rows = TRUE, cluster_columns = TRUE, show_column_names = F, #col = col_fun,
show_row_dend = F, show_column_dend = F, # do not show dendrogram
row_names_gp = gpar(fontsize = 8), column_title="Phenotype Corr (RankZ, Pearson)", column_title_gp = gpar(fontsize = 8),
name="Corr")
pheno.order <- row_order(ht)Warning: The heatmap has not been initialized. You might have different results
if you repeatedly execute this function, e.g. when row_km/column_km was
set. It is more suggested to do as `ht = draw(ht); row_order(ht)`.#draw(ht)
BC.cor.rank.fig <- BC.cor.rank.fig[pheno.order,pheno.order]
ht1 = Heatmap(BC.cor.rank.fig, cluster_rows = FALSE, cluster_columns = FALSE, show_column_names = F, #col = col_fun,
show_row_dend = F, show_column_dend = F, # do not show dendrogram
row_names_gp = gpar(fontsize = 8), column_title="Phenotype Corr (RankZ, Pearson)", column_title_gp = gpar(fontsize = 8),
name="Corr")
BC.cor.fig <- BC.cor.fig[pheno.order,pheno.order]
ht2 = Heatmap(BC.cor.fig, cluster_rows = FALSE, cluster_columns = FALSE, show_column_names = F, #col = col_fun,
row_names_gp = gpar(fontsize = 8), column_title="Phenotype Corr (Spearman)", column_title_gp = gpar(fontsize = 8),
name="Corr")
BC.cor.combat.fig <- BC.cor.combat.fig[pheno.order,pheno.order]
ht3 = Heatmap(BC.cor.combat.fig, cluster_rows = FALSE, cluster_columns = FALSE, show_column_names = F, #col = col_fun,
row_names_gp = gpar(fontsize = 8), column_title="Phenotype Corr (Combat, Pearson)", column_title_gp = gpar(fontsize = 8),
name="Corr")
BC.zcor.fig <- BC.zcor.fig[pheno.order,pheno.order]
ht4 = Heatmap(BC.zcor.fig, cluster_rows = FALSE, cluster_columns = FALSE, show_column_names = F, #col = col_fun,
row_names_gp = gpar(fontsize = 8), column_title="Genetic Corr (Pearson)", column_title_gp = gpar(fontsize = 8),
name="Corr"
)
draw(ht1+ht2+ht3+ht4)Warning: Heatmap/annotation names are duplicated: CorrWarning: Heatmap/annotation names are duplicated: Corr, CorrWarning: Heatmap/annotation names are duplicated: Corr, Corr, Corr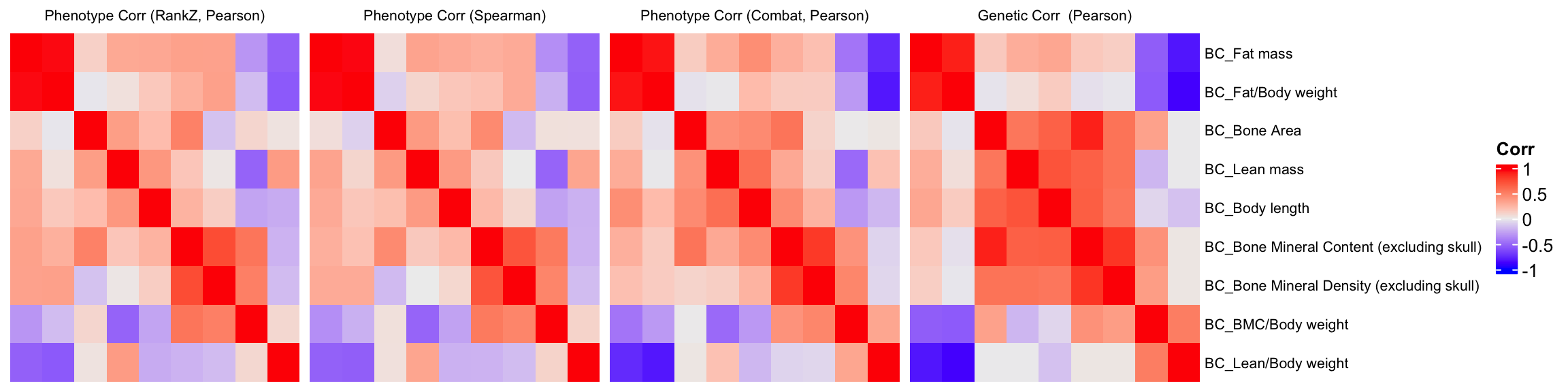
| Version | Author | Date |
|---|---|---|
| 7685a09 | statsleelab | 2023-01-10 |
png(file="docs/figure/figures.Rmd/cors_BC.png", width=800, height=250)
draw(ht1+ht2+ht3+ht4)Warning: Heatmap/annotation names are duplicated: CorrWarning: Heatmap/annotation names are duplicated: Corr, CorrWarning: Heatmap/annotation names are duplicated: Corr, Corr, Corrdev.off()quartz_off_screen
2 Test of the correlation between genetic correlation matrices
We use the Mantel’s test for testing the correlation between two distance matrices.
####################
# Use Mantel test
# https://stats.idre.ucla.edu/r/faq/how-can-i-perform-a-mantel-test-in-r/
# install.packages("ade4")
library(ade4)
to.upper<-function(X) X[upper.tri(X,diag=FALSE)]
a1 <- to.upper(BC.cor.fig)
a2 <- to.upper(BC.cor.rank.fig)
a3 <- to.upper(BC.cor.combat.fig)
a4 <- to.upper(BC.zcor.fig)
plot(a4, a1)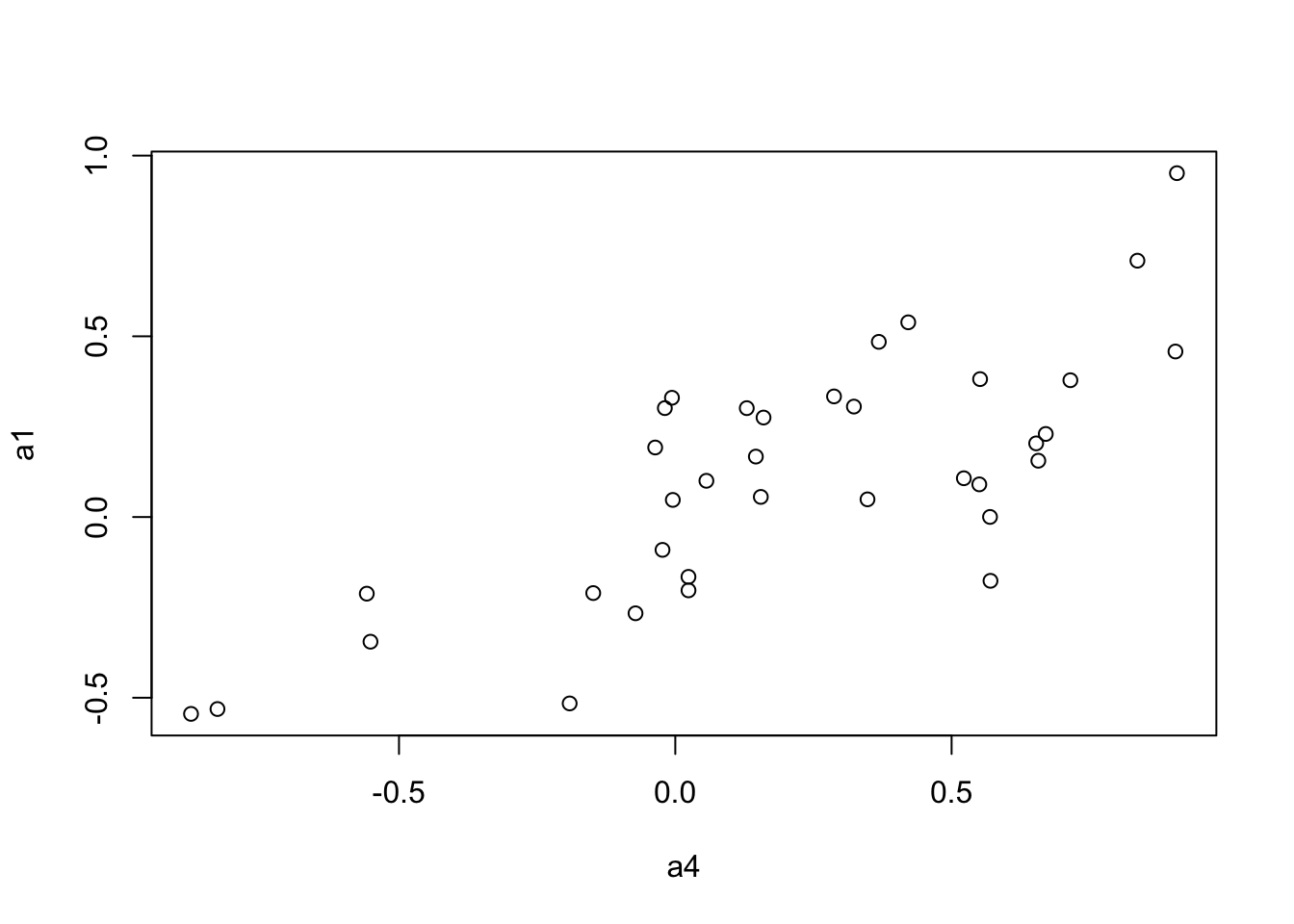
| Version | Author | Date |
|---|---|---|
| 7685a09 | statsleelab | 2023-01-10 |
plot(a4, a2)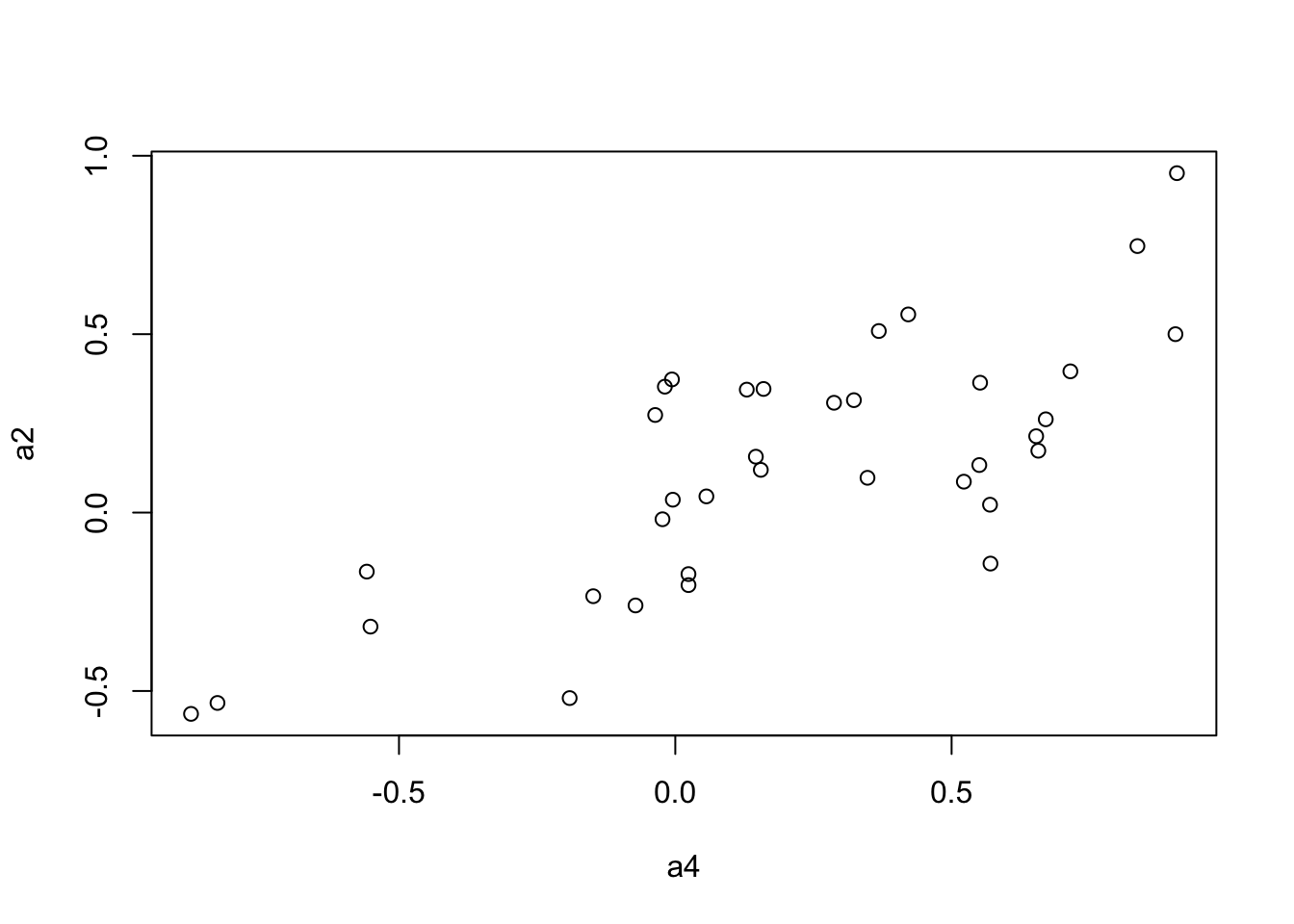
| Version | Author | Date |
|---|---|---|
| 7685a09 | statsleelab | 2023-01-10 |
plot(a4, a3)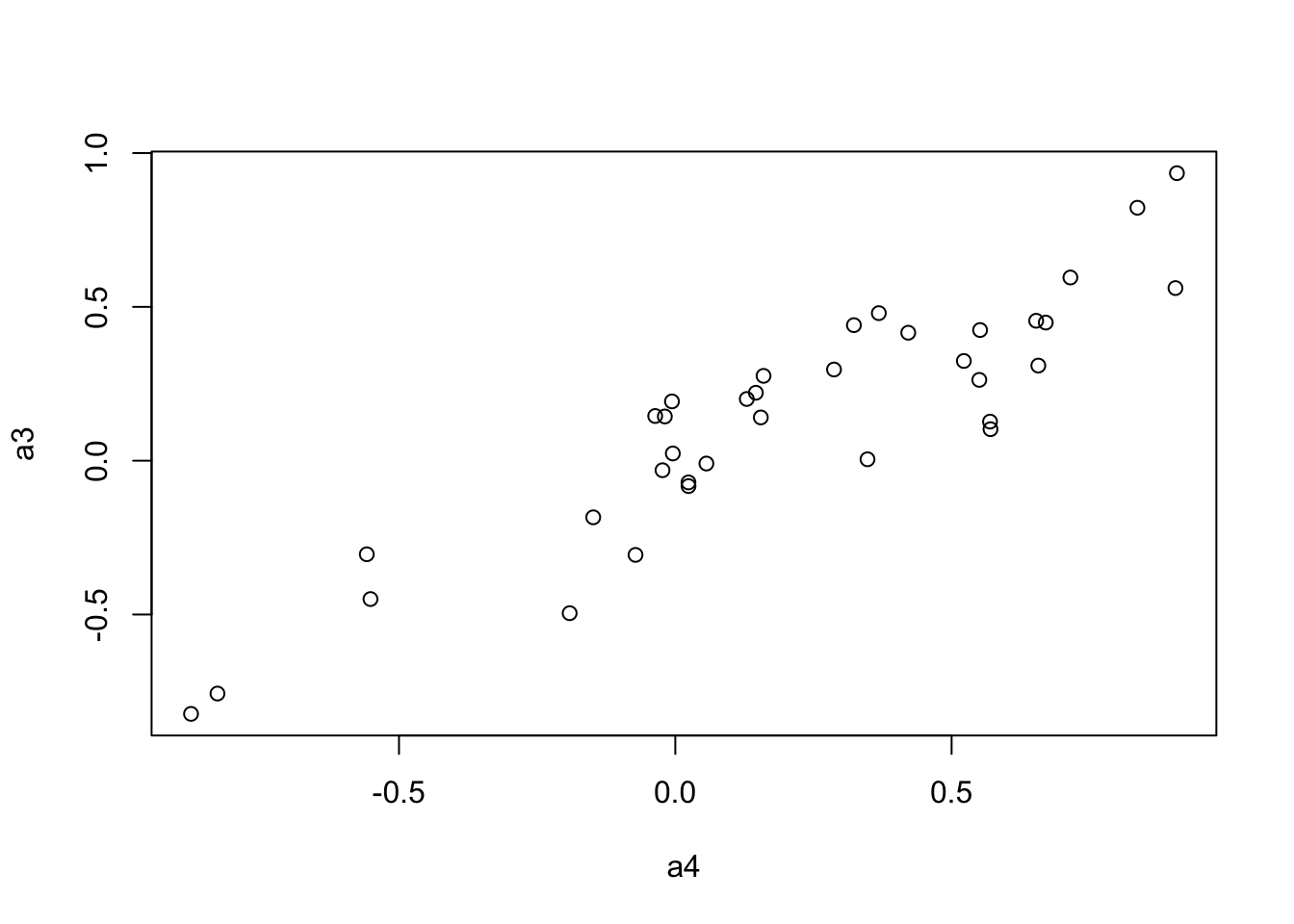
| Version | Author | Date |
|---|---|---|
| 7685a09 | statsleelab | 2023-01-10 |
mantel.rtest(as.dist(1-BC.cor.fig), as.dist(1-BC.zcor.fig), nrepet = 9999) #nrepet = number of permutationsMonte-Carlo test
Call: mantelnoneuclid(m1 = m1, m2 = m2, nrepet = nrepet)
Observation: 0.7588338
Based on 9999 replicates
Simulated p-value: 1e-04
Alternative hypothesis: greater
Std.Obs Expectation Variance
4.2777904374 0.0007922433 0.0314012571 mantel.rtest(as.dist(1-BC.cor.rank.fig), as.dist(1-BC.zcor.fig), nrepet = 9999)Monte-Carlo test
Call: mantelnoneuclid(m1 = m1, m2 = m2, nrepet = nrepet)
Observation: 0.7527398
Based on 9999 replicates
Simulated p-value: 1e-04
Alternative hypothesis: greater
Std.Obs Expectation Variance
4.138201139 -0.001075877 0.033182353 mantel.rtest(as.dist(1-BC.cor.combat.fig), as.dist(1-BC.zcor.fig), nrepet = 9999)Monte-Carlo test
Call: mantelnoneuclid(m1 = m1, m2 = m2, nrepet = nrepet)
Observation: 0.9076102
Based on 9999 replicates
Simulated p-value: 1e-04
Alternative hypothesis: greater
Std.Obs Expectation Variance
4.7809904425 -0.0005658379 0.0360831394 Test KOMPUTE imputation algorithm
Load KOMPUTE package
if(!"kompute" %in% rownames(installed.packages())){
library(devtools)
devtools::install_github("dleelab/kompute")
}
library(kompute)Simulation study - imputed vs measured
We randomly select measured gene-phenotype association z-scores, mask those, impute them using KOMPUTE method. Then we compare the imputed z-scores to the measured ones.
zmat <-t(BC.zmat)
dim(zmat)[1] 9 4412## filter genes with na < 10
zmat0 <- is.na(zmat)
num.na<-colSums(zmat0)
summary(num.na) Min. 1st Qu. Median Mean 3rd Qu. Max.
0.000 0.000 1.000 1.708 3.000 8.000 zmat <- zmat[,num.na<10]
dim(zmat)[1] 9 4412#pheno.cor <- BC.cor.fig
#pheno.cor <- BC.cor.rank.fig
pheno.cor <- BC.cor.combat.fig
#pheno.cor <- BC.zcor.fig
zmat <- zmat[rownames(pheno.cor),,drop=FALSE]
rownames(zmat)[1] "BC_Fat mass"
[2] "BC_Fat/Body weight"
[3] "BC_Bone Area"
[4] "BC_Lean mass"
[5] "BC_Body length"
[6] "BC_Bone Mineral Content (excluding skull)"
[7] "BC_Bone Mineral Density (excluding skull)"
[8] "BC_BMC/Body weight"
[9] "BC_Lean/Body weight" rownames(pheno.cor)[1] "BC_Fat mass"
[2] "BC_Fat/Body weight"
[3] "BC_Bone Area"
[4] "BC_Lean mass"
[5] "BC_Body length"
[6] "BC_Bone Mineral Content (excluding skull)"
[7] "BC_Bone Mineral Density (excluding skull)"
[8] "BC_BMC/Body weight"
[9] "BC_Lean/Body weight" colnames(pheno.cor)[1] "BC_Fat mass"
[2] "BC_Fat/Body weight"
[3] "BC_Bone Area"
[4] "BC_Lean mass"
[5] "BC_Body length"
[6] "BC_Bone Mineral Content (excluding skull)"
[7] "BC_Bone Mineral Density (excluding skull)"
[8] "BC_BMC/Body weight"
[9] "BC_Lean/Body weight" npheno <- nrow(zmat)
## percentage of missing Z-scores in the original data
100*sum(is.na(zmat))/(nrow(zmat)*ncol(zmat)) # 21%[1] 18.97351nimp <- 2000 # # of missing/imputed Z-scores
set.seed(1111)
## find index of all measured zscores
all.i <- 1:(nrow(zmat)*ncol(zmat))
measured <- as.vector(!is.na(as.matrix(zmat)))
measured.i <- all.i[measured]
## mask 2000 measured z-scores
mask.i <- sort(sample(measured.i, nimp))
org.z = as.matrix(zmat)[mask.i]
zvec <- as.vector(as.matrix(zmat))
zvec[mask.i] <- NA
zmat.imp <- matrix(zvec, nrow=npheno)
rownames(zmat.imp) <- rownames(zmat)Run KOMPUTE method
kompute.res <- kompute(zmat.imp, pheno.cor, 0.01)
KOMPute running...# of genes: 4412# of phenotypes: 9# of imputed Z-scores: 9534# measured vs imputed
length(org.z)[1] 2000imp.z <- as.matrix(kompute.res$zmat)[mask.i]
imp.info <- as.matrix(kompute.res$infomat)[mask.i]
plot(imp.z, org.z)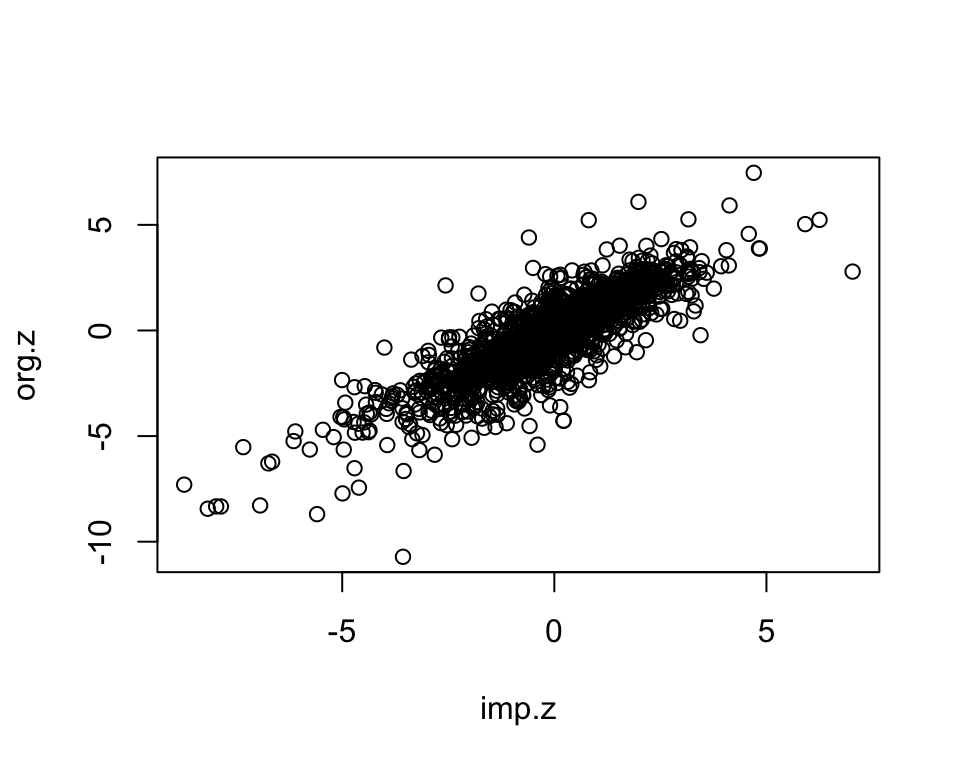
| Version | Author | Date |
|---|---|---|
| 7685a09 | statsleelab | 2023-01-10 |
imp <- data.frame(org.z=org.z, imp.z=imp.z, info=imp.info)
dim(imp)[1] 2000 3imp <- imp[complete.cases(imp),]
imp <- subset(imp, info>=0 & info <= 1)
dim(imp)[1] 1995 3cor.val <- round(cor(imp$imp.z, imp$org.z), digits=3)
cor.val[1] 0.852plot(imp$imp.z, imp$org.z)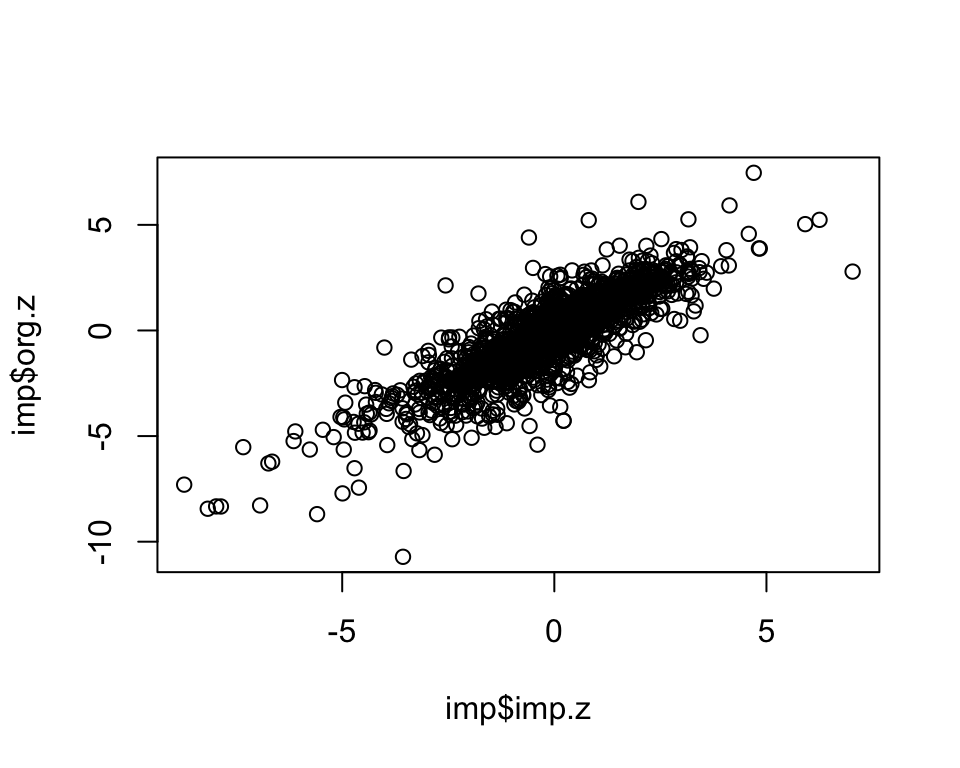
| Version | Author | Date |
|---|---|---|
| 7685a09 | statsleelab | 2023-01-10 |
info.cutoff <- 0.8
imp.sub <- subset(imp, info>info.cutoff)
dim(imp.sub)[1] 1174 3summary(imp.sub$imp.z) Min. 1st Qu. Median Mean 3rd Qu. Max.
-8.72493 -1.12303 -0.08752 -0.21512 0.80615 7.03123 summary(imp.sub$info) Min. 1st Qu. Median Mean 3rd Qu. Max.
0.8052 0.8516 0.8828 0.8989 0.9565 0.9715 cor.val <- round(cor(imp.sub$imp.z, imp.sub$org.z), digits=3)
cor.val[1] 0.904g <- ggplot(imp.sub, aes(x=imp.z, y=org.z, col=info)) +
geom_point() +
labs(title=paste0("IMPC Behavior Data (BC), Info>", info.cutoff, ", Cor=",cor.val),
x="Imputed Z-scores", y = "Measured Z-scores", col="Info") +
theme_minimal()
g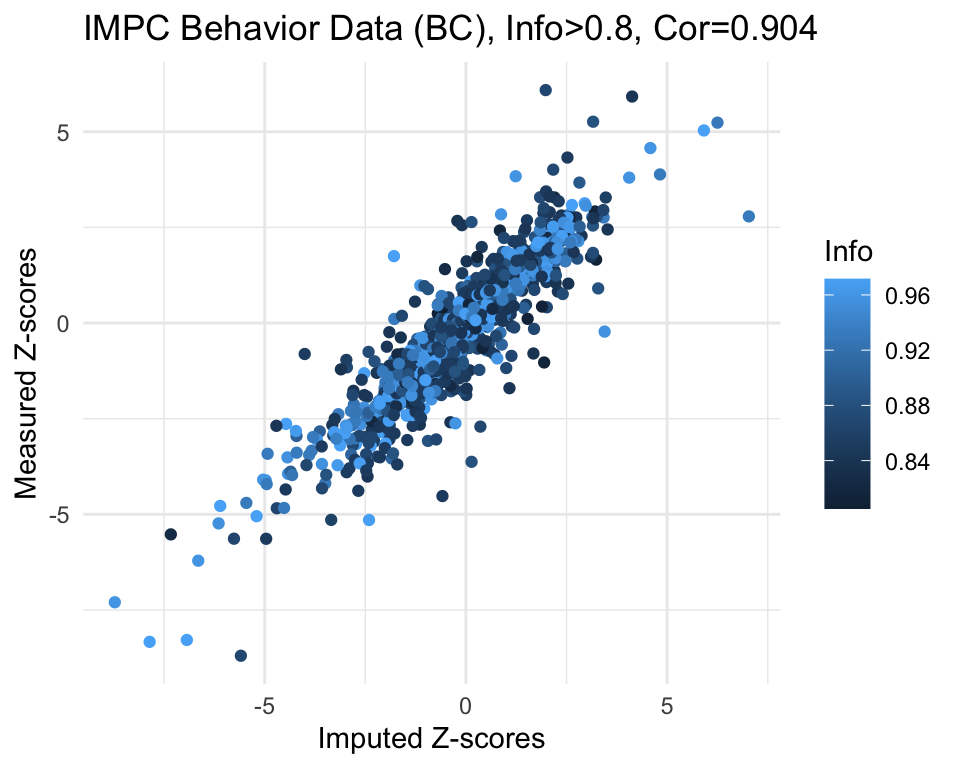
| Version | Author | Date |
|---|---|---|
| 7685a09 | statsleelab | 2023-01-10 |
# save plot
png(file="docs/figure/figures.Rmd/sim_results_BC.png", width=600, height=350)
g
dev.off()quartz_off_screen
2
sessionInfo()R version 4.2.1 (2022-06-23)
Platform: x86_64-apple-darwin17.0 (64-bit)
Running under: macOS Catalina 10.15.7
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] grid stats graphics grDevices utils datasets methods
[8] base
other attached packages:
[1] kompute_0.1.0 ade4_1.7-20 sva_3.44.0
[4] BiocParallel_1.30.3 genefilter_1.78.0 mgcv_1.8-40
[7] nlme_3.1-158 lme4_1.1-31 Matrix_1.5-1
[10] RNOmni_1.0.1 ComplexHeatmap_2.12.1 circlize_0.4.15
[13] RColorBrewer_1.1-3 tidyr_1.2.0 ggplot2_3.4.1
[16] reshape2_1.4.4 dplyr_1.0.9 data.table_1.14.2
[19] workflowr_1.7.0.1
loaded via a namespace (and not attached):
[1] minqa_1.2.5 colorspace_2.1-0 rjson_0.2.21
[4] rprojroot_2.0.3 XVector_0.36.0 GlobalOptions_0.1.2
[7] fs_1.5.2 clue_0.3-62 rstudioapi_0.13
[10] farver_2.1.1 bit64_4.0.5 AnnotationDbi_1.58.0
[13] fansi_1.0.4 codetools_0.2-18 splines_4.2.1
[16] doParallel_1.0.17 cachem_1.0.6 knitr_1.39
[19] jsonlite_1.8.0 nloptr_2.0.3 annotate_1.74.0
[22] cluster_2.1.3 png_0.1-8 compiler_4.2.1
[25] httr_1.4.3 assertthat_0.2.1 fastmap_1.1.0
[28] limma_3.52.4 cli_3.6.0 later_1.3.0
[31] htmltools_0.5.3 tools_4.2.1 gtable_0.3.1
[34] glue_1.6.2 GenomeInfoDbData_1.2.8 Rcpp_1.0.10
[37] Biobase_2.56.0 jquerylib_0.1.4 vctrs_0.5.2
[40] Biostrings_2.64.0 iterators_1.0.14 xfun_0.31
[43] stringr_1.4.0 ps_1.7.1 lifecycle_1.0.3
[46] XML_3.99-0.10 edgeR_3.38.4 getPass_0.2-2
[49] MASS_7.3-58.1 zlibbioc_1.42.0 scales_1.2.1
[52] promises_1.2.0.1 parallel_4.2.1 yaml_2.3.5
[55] memoise_2.0.1 sass_0.4.2 stringi_1.7.8
[58] RSQLite_2.2.15 highr_0.9 S4Vectors_0.34.0
[61] foreach_1.5.2 BiocGenerics_0.42.0 boot_1.3-28
[64] shape_1.4.6 GenomeInfoDb_1.32.3 rlang_1.0.6
[67] pkgconfig_2.0.3 matrixStats_0.62.0 bitops_1.0-7
[70] evaluate_0.16 lattice_0.20-45 purrr_0.3.4
[73] labeling_0.4.2 bit_4.0.4 processx_3.7.0
[76] tidyselect_1.2.0 plyr_1.8.7 magrittr_2.0.3
[79] R6_2.5.1 IRanges_2.30.0 generics_0.1.3
[82] DBI_1.1.3 pillar_1.8.1 whisker_0.4
[85] withr_2.5.0 survival_3.3-1 KEGGREST_1.36.3
[88] RCurl_1.98-1.8 tibble_3.1.8 crayon_1.5.1
[91] utf8_1.2.3 rmarkdown_2.14 GetoptLong_1.0.5
[94] locfit_1.5-9.6 blob_1.2.3 callr_3.7.1
[97] git2r_0.30.1 digest_0.6.29 xtable_1.8-4
[100] httpuv_1.6.5 stats4_4.2.1 munsell_0.5.0
[103] bslib_0.4.0